GCPBayes Pipeline
Published in Physica A, 2008
This paper is about using probabilistic cellular automata for calculation of the critical points for the anisotropic two-layer Ising and Potts models.
Citation: Y. Asgari, M. Ghaemi, (2008). "Obtaining the critical point and shift exponent for the anisotropic two-layer Ising and Potts models: Cellular Automata approach" Physica A: Statistical Mechanics and its Applications. Vol.387, No.8-9, Page:1937-1946 https://www.sciencedirect.com/science/article/abs/pii/S0378437107012265
Published in IRANIAN JOURNAL OF CHEMISTRY AND CHEMICAL ENGINEERING, 2011
This paper is about using the Cellular Automata method to simulate the pattern formation of the FitzHugh-Nagumo (FHN) model.
Citation: Y. Asgari, M. Ghaemi, M.G. Mahjani, (2011). "Pattern formation of the FitzHugh-Nagumo model: cellular automata approach" IRANIAN JOURNAL OF CHEMISTRY AND CHEMICAL ENGINEERING. Vol.30, No.1, Page:135-142 https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0079397
Published in PloS One, 2013
This paper is about investigation of particular characteristics in cancer metabolic networks.
Citation: Y. Asgari, A. Salehzadeh-Yazdi, F. Schreiber, A. Masoudi-Nejad, (2013). "Controllability in Cancer Metabolic Networks According to Drug Targets as Driver Nodes" PloS One. Vol.8, No.11, Page:e79397 https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0079397
Published in Seminars in Cancer Biology, 2015
This paper is about advances in topological and constraint-based methods for analysis of cancer as a metabolic disease.
Citation: A. Masoudi-Nejad, Y. Asgari, (2015). "Metabolic Cancer Biology: Structural-based analysis of cancer as a metabolic disease, new sights and opportunities for disease treatment" Seminars in Cancer Biology. Vol.30, Page:21-29 https://www.sciencedirect.com/science/article/abs/pii/S1044579X14000236
Published in Genomics, 2015
Using reconstructed tissue-specific metabolic networks for cancer and normal cells, this paper explores whether the Warburg effect is a consequence of metabolic adaptation in cancer cells.
Citation: Y. Asgari, Z. Zabihinpour, A. Salehzadeh-Yazdi, F. Schreiber, A. Masoudi-Nejad, (2015). "Alterations in cancer cells metabolism: the Warburg effect and metabolic adaptation" Genomics. Vol.105, Page:275-281 https://www.sciencedirect.com/science/article/pii/S088875431500049X
Published in Current Bioinformatics, 2018
We have developed a MATLAB/Octave toolbox, which extends the COBRA framework and provides functions to prepare different metabolite- and enzyme-centric networks that could also be imported in other structural analysis softwares, such as Cytoscape for structural analyses.
Citation: Y. Asgari, Z. Zabihinpour, A. Masoudi-Nejad, (2018). "SCAN-Toolbox: Structural COBRA Add-oN (SCAN) for Analysing Large Metabolic Networks" Genomics. Vol.13, No.1, Page:100-107 https://www.ingentaconnect.com/content/ben/cbio/2018/00000013/00000001/art00015
Published in Integrative Biology, 2018
We have developed a complementary approach (integration of co-expression networks and genome-scale metabolic models) for lung and prostate, under normal and cancer conditions which could lead to proposing novel biomarkers.
Citation: Y. Asgari, P. Khosravi, Z. Zabihinpour, M. Habibi, (2018). "Exploring candidate biomarkers for lung and prostate cancers using gene expression and flux variability analysis" Integrative Biology. Vol.10, No.113, Page:113-120 https://academic.oup.com/ib/article-abstract/10/2/113/5115777
Published in MATCH Communications in Mathematical and in Computer Chemistry, 2019
In this paper, we have used deterministic and stochastic approaches to show that the average and the standard deviation of ligand binding capacity indeed differ between positive and negative cooperativity in some protein multimers. Considering standard deviation as a measure of noise in the system, our results demonstrated that this source of noise enables the system to adapt in a fluctuating environment and could be subject to selection during evolution.
Citation: M. Taheri, K. Nouri, Y. Asgari, Z. Zabihinpour, M. Sadeghi, (2019). "The Significance of Noise in the Evolution of Negative and Positive Cooperativity in Protein Complexes" MATCH Communications in Mathematical and in Computer Chemistry. Vol.81, No.1, Page:177-192 https://match.pmf.kg.ac.rs/electronic_versions/Match81/n1/match81n1_177-192.pdf
Published in Scientific Reports, 2020
In this paper, we have created the iMM1865 model as an orthology-based reconstruction of a genome-scale metabolic (GSM) model for Mus musculus based on the flux-consistent version of the human metabolic network, Recon3D.
Citation: S. Khodaee, Y. Asgari, M. Totochi, M.H. Karimi-Jafari, (2020). "iMM1865: A New Reconstruction of Mouse Genome-Scale Metabolic Model" Scientific Reports. Vol 10, No.1, Page:1-13 https://www.nature.com/articles/s41598-020-63235-w
Published in biorxiv, 2020
We identified novel neurodevelopment disorders-associated ncRNAs that influence genomic enhancers during neurodevelopment by integrating genome-wide association study (GWAS), copy number variation (CNV), and expression quantitative trait loci (eQTL) data.
Citation: Y. Asgari, J. Ik-Tsen Heng, N. Lovell, A. Forrest, H. Alinejad-Rokny, (2020). "Evidence for enhancer noncoding RNAs (enhancer-ncRNAs) with gene regulatory functions relevant to neurodevelopmental disorders" biorxiv. https://www.biorxiv.org/content/10.1101/2020.05.16.087395v2.abstract
Published in SN Applied Sciences, 2020
In this paper, we have explored an assumption in which there exists a shift in playing strategy of enzymes from normal to a cancer condition by investigation of pattern alterations in the metabolic subsystem of four cancer cell types in four cancer stages using flux calculations and Gene Ontology analyses.
Citation: Y. Asgari, P. Khosravi, (2020). "Flux variability analysis reveals a tragedy of commons in cancer cells" SN Applied Sciences. 2, 1966 https://link.springer.com/article/10.1007/s42452-020-03762-3
Published in Scientific Reports, 2020
In this paper, we have used a systems biology approach to find more suitable targets in order to restore sodium-iodide symporter (NIS) expression in ATC cells. We have built a simplified protein interaction network including transcription factors and proteins involved in signaling pathways which regulate NIS expression.
Citation: H. Rakhsh-Khorshid1, H. Samimi, S. Torabi, S.M. Sajjadi-Jazi, H. Samadi, F. Ghafouri, Y. Asgari, V. Haghpanah, (2020). "Network analysis reveals essential proteins that regulate sodium-iodide symporter expression in anaplastic thyroid carcinoma" Scientific Reports. Vol 10, 21440 https://link.springer.com/content/pdf/10.1038/s41598-020-78574-x.pdf
Published in PloS One, 2022
In this paper, we have studied the spread of Ebola virus in lymph nodes using a simplified Cellular Automata model with only four parameters.
Citation: M. Ghaemi, M. Shojafar, Z. Zabihinpour, Y. Asgari, (2022). "On the possibility of oscillating in the Ebola virus dynamics and investigating the effect of the lifetime of T lymphocytes" PloS One. 17(3): e0265065 https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0265065
Published in NAR Genomics and Bioinformatics, 2023
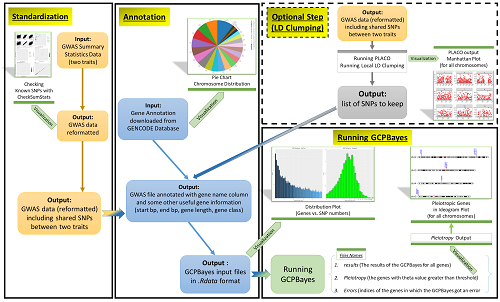
In this paper, we designed a user-friendly pipeline to perform cross-phenotype gene-set analysis between two traits using GCPBayes, a method developed by our team.
Citation: Y. Asgari, PE. Sugier, T. Baghfalaki, E. Lucotte, M. Karimi, M. Sedki, A. Ngo, B. Liquet, T. Truong, (2023). "GCPBayes pipeline: a tool for exploring pleiotropy at the gene level" NAR Genomics and Bioinformatics. 5(3): lqad065 https://academic.oup.com/nargab/article/5/3/lqad065/7219410
Published in R Journal, 2023
In this paper, we developed an R package to study cross-phenotypes associations (or pleiotropy) at the Gene-level, based on summary statistics data from genome-wide association studies (GWAS).
Citation: T. Baghfalaki, PE. Sugier, Y. Asgari, T. Truong, B. Liquet, (2023). "GCPBayes: An R package for studying Cross-Phenotype Genetic Associations with Group-level Bayesian Meta-Analysis" R Journal. 15(1): 122-141 https://rjournal.github.io/articles/RJ-2023-028/
Published in Human Molecular Genetics, 2024
In this study, we investigated common genetic risk factors between breast cancer and thyroid cancer or thyroid hormones levels.
Citation: E.A. Lucotte, Y. Asgari, P.E. Sugier, M. Karimi, C. Domenighetti, F. Lesueur, A. Boland-Augé, E. Ostroumova, F. de Vathaire, M. Zidane, P. Guénel, J.F. Deleuze, M.C.e Boutron-Ruault, G. Severi, B. Liquet, T. Truong, (2024). "Investigation of common genetic risk factors between thyroid traits and breast cancer" Human Molecular Genetics. 33(1), 38–47 https://academic.oup.com/hmg/article/33/1/38/7280729
Published in Biostatistics, 2025
In this study, we proposed a new Meta-analysis model adapted for Pleiotropy Selection with Group structure.
Citation: P.E. Sugier, Y. Asgari, M. Sedki, T. Truong, B. Liquet, (2025). "Meta-analysis models with group structure for pleiotropy detection at gene and variant level using summary statistics from multiple datasets. 26(1), kxaf037 https://academic.oup.com/biostatistics/article-abstract/26/1/kxaf037/8324642?redirectedFrom=fulltext
Published:
Published:
In this Poster, we illustrated a new pipeline to use GCPBayes to detect pleiotropy between 2 traits using GWAS summary statistics data as inputs. To show its applicability, we used this pipeline it to detect common genetic risk factors between breast cancer and ovarian cancer using data from two large consortia. We compared the highlighted genes to available findings from the literature.
Published:
My talk was about a new pipeline we developed for analysis of pleiotropy at gene-level using summary statistics GWAS data.
Published:
My talk was about a new pipeline we developed for analysis of pleiotropy at gene-level using summary statistics GWAS data.
Published:
My talk was about a new pipeline we developed for analysis of pleiotropy at gene-level using summary statistics GWAS data.
Published:
In this Poster, we illustrated the use of GCPBayes Pipeline (our developed pipeline) by exploring pleiotropic genes between breast cancer and thyroid cancer.
Published:
In this Poster, we illustrated the results of our study to investigate potential pleiotropic genes and pathways between BC and three thyroid traits (thyroid cancer, TSH levels, and FT4 levels) using a gene-set approach.
Published:
My talk was about the findings from our paper on investigation of pleiotropy between 3 different Thyroid traits and Breast Cancer Risk using a gene-based level approach.
Published:
My talk was about our recent findings on Epigenetic Age Acceleration and Breast Cancer Risk within the French E3N-Generations Cohort.
Graduate Course, University of Tehran, Department of Bioinformatics, 2016
Graduate Course, Tehran University of Medical Sciences, Department of Medical Biotechnology, 2018
Graduate Course, Tehran University of Medical Sciences, Department of Medical Biotechnology, 2019
You could find “Research Method” course information (overview, Textbooks, Lecture Schedule and details) in the following page: “Research Method Course Details in my GitHub Page”
Graduate Course, Tehran University of Medical Sciences, Department of Medical Biotechnology, 2019
You could find “Systems Biology for Tissue Engineering” course information (overview, Textbooks, Lecture Schedule and details) in the following page: “Systems Biology for Tissue Engineering Course Details in my GitHub Page”
Graduate Course, Tehran University of Medical Sciences, Department of Medical Biotechnology, 2020
You could find “Medical Informatics” course information (overview, Textbooks, Lecture Schedule and details) in the following page: “Medical Informatics Course Details in my GitHub Page”
Graduate Course, Tehran University of Medical Sciences, Department of Medical Biotechnology, 2020
You could find “Bioinformatics” course information (overview, Textbooks, Lecture Schedule and details) in the following page: “Bioinformatics Course Details in my GitHub Page”
Graduate Course, Tehran University of Medical Sciences, Department of Medical Biotechnology, 2020
You could find “Protein Engineering” course information (overview, Textbooks, Lecture Schedule and details) in the following page: “Protein Engineering Course Details in my GitHub Page”
Graduate Course, Tehran University of Medical Sciences, Department of Medical Biotechnology, 2020
You could find “Systems Biology” course information (overview, Textbooks, Lecture Schedule and details) in the following page: “Systems Biology Course Details in my GitHub Page”